Current and past research:
Probabilistic modelling of single-cell genomics data
Current research interests in this area include creating statistical machine learning models and scalable inference paradigms to understand cell type composition, interactions, and signatures from single-cell and spatially-resolved 'omics data with a focus on the tumour microenvironment.
Research works:
We created CellAssign, a novel statistical model implemented in Google's Tensorflow to assign cells to known tumour microenvironment cell types from single-cell RNA-sequencing data, across large patient cohorts while controlling for sample and technical effects.

CloneAlign assigns cells as measured with single-cell RNA-seq to mutational cancer clones defined by copy number profiles by probabilistically mapping RNA-seq to clone-specific copy number profiles using reparametrization gradient variational inference.
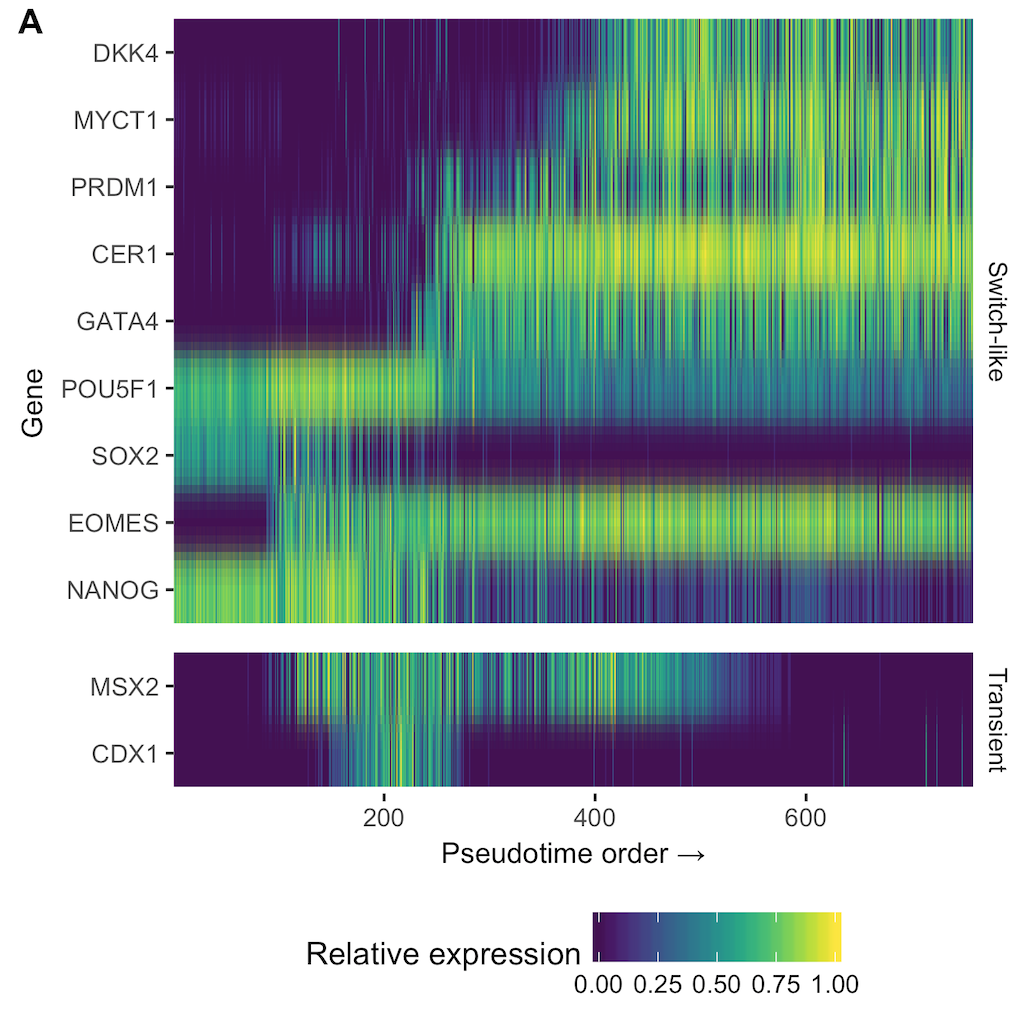
We have created a number of methods to understand single-cell trajectories from a probabilistic perspective, in particular (i) while controlling for heterogeneous genetic or phenotypic backgrounds, (ii) directly from marker genes allowing interpretable inference of pseudotimes, (iii) probabilistic inference of bifurcations.
Campbell and Yau (Nature Communications)
Disease genomics & cancer
Current research interests include how the tumour microenvironment modulates both tumour evolution and phenotypic state, as well as deriving novel progression scores and signatures from high dimensional clinical, imaging, and 'omics data.
Research works:We performed single-cell RNA-sequencing of a large number of solid tumour types (patient derived xenografts, cell lines, primary tumour samples) in breast and ovarian cancers to uncover the effect of tissue dissociation using collagenase at 37C to alternative enzymes at lower temperatures. We found a marked upregulation of stress pathway related genes.
We performed single-cell RNA-sequencing of iPSC-derived dopaminergic neurons from Parkinson's patients and controls to reconstruct disease progression at single-cell level. Through differential expression analysis we uncovered mis-localization of the protein HDAC4 being implicated in Parkinson's progression.
Probabilistic machine learning
Current research interests in this area include statistical machine learning methods for understanding structure and outcome prediction calibration from large heterogeneous datasets such as EHRs in the presence of high levels of missing data.
Research works: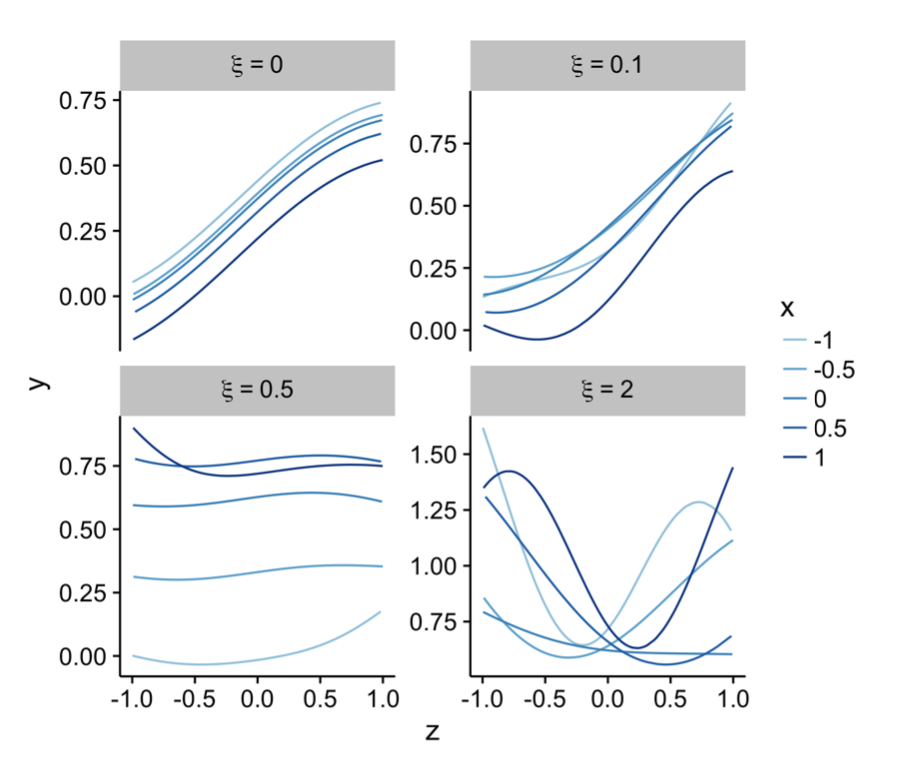
Covariate Gaussian Process Latent Variable Models (C-GPLVMs) are a novel type of probabilistic latent variable model similar to a GPLVM except the functional form of the outputs wrt the latent space is modified by an additional set of covariates.